Data generation is very important and useful for developing and testing new database algorithms. (Generated data is usually referred to as synthetic data.) First, it's easier to control the statistical characteristics of synthetic data which can predict the correct result theoretically and hence help testing the new algorithms. Second, it's not always easy to get interesting and easy-to-use realistic database. For the same reason, data generation plays an important role in our project as well, especially since it's very difficult to get a medical database containing real-world sensitive data because we would have to go through some clearance process first.
The data generation in this project contains 3 parts: Taxonomy generation, tuple-set generation and association rule generation.
For the Taxonomy![]() generation we have two parameters to control the generation of the
hierarchy tree: the maximum depth of the hierarchy tree , and
the maximum node fanout . The entire hierarchy tree stands
for one categorical attribute. Each leaf in the hierarchy tree forms
one binary attribute and corresponds to one value of the categorical
attribute. Figure 1 describes this point, please
note that only the leaves have an attribute number. We call the group
of binary attributes corresponding to one categorical attribute an
attribute group. The total number of attribute groups
is therefore the number of attributes in our medical
database. We chose the values and .
Figure 2 shows another tree that was generated. On
average one attribute group has around 20 leaves. The total number
of binary attributes generated varies. talk about
max size, etc.
generation we have two parameters to control the generation of the
hierarchy tree: the maximum depth of the hierarchy tree , and
the maximum node fanout . The entire hierarchy tree stands
for one categorical attribute. Each leaf in the hierarchy tree forms
one binary attribute and corresponds to one value of the categorical
attribute. Figure 1 describes this point, please
note that only the leaves have an attribute number. We call the group
of binary attributes corresponding to one categorical attribute an
attribute group. The total number of attribute groups
is therefore the number of attributes in our medical
database. We chose the values and .
Figure 2 shows another tree that was generated. On
average one attribute group has around 20 leaves. The total number
of binary attributes generated varies. talk about
max size, etc.
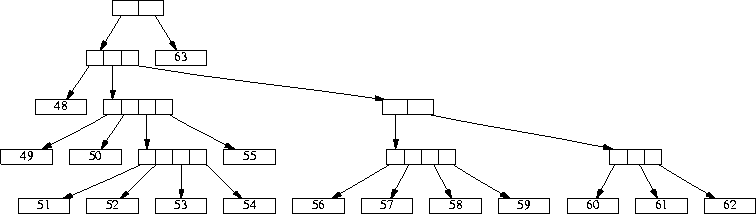
Figure 1: A sample hierarchy tree
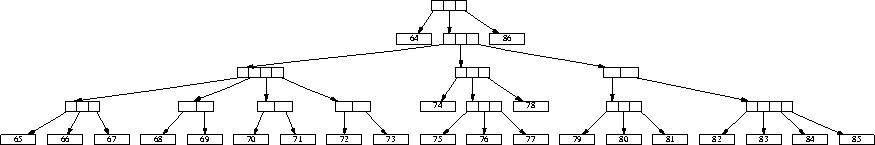
Figure 2: Another sample hierarchy tree
Once we have generated the taxonomy, we can generate the association rules and the tuple sets. The parameters for the association rule are the number of association rules to generate and the maximum number of literals per association rule . As mentioned earlier, our association rules are of the form (we designate as the length of the association rule). Ideally our association rules do not partition the positive tuples into disjunct sets, but any positive tuple can be a match for any subset of rules. In our opinion, this models the reality of a medical database the closest.
Conceptually, the association rules are generated first, along with a desired support and confidence parameter. The tuples are then generated accordingly to these parameters. This sequence however, leads to complicated computations of which elements in the positive and negative tuple set need to match which association rules to satisfy the support and confidence parameter of each rule. We have used the following trick to simplify the generation. The positive and negative tuples are generated first in a totally random manner. We keep the number of true attributes small to model reality closer (i.e. it does not make sense for a person to take 10 out of 100 medications, the actual number of distinct medications taken is usually small). In addition we did not want to introduce unintended associations. If we set a high number of binary attributes to true, the probability is high that the data would contain an unintended, hidden association rule. Given the total number of binary attributes , we set to true, where is a normal distributed random variable with average and . We were satisfied with the resulting number of true attributes. discuss how it changes and how many total attributes we targeted
The next step then was to generate the association rules. Our objective was to generate rules which matched only few of the randomly generated tuples. The intuition behind this scheme is that if an association rule matches many randomly generated tuples, then it is too general. So we chose a threshold 1% and if the number of matching tuples in the randomly generated tuples passed it, the association rule was discarded. We then assigned to every found association rule a support parameter in the interval with standing for the uniform distribution. This support only determines matching tuples in the positive tuple set. We then generated the confidence parameter, which we set to be higher than 99%. This parameter determined how many tuples in the negative tuple set were to be set. Again we used a uniform distribution to compute the actual number .
Knowing the number of desired matches in each the positive and negative tuple set for each association rule, it was a simple task to generate the data. For each association rule, we choose as many tuples as needed in a random manner and set the attributes needed to transform the tuple into a matching tuple. The method used can be described best by looking at an association rule as a set of nodes in our forest of hierarchy trees -- each literal in the rule corresponds to exactly one node in the tree. Figure 3 shows a tree with the node numbers printed. The number of binary attributes per literal that were actually set were determined after a uniform distribution where stands for the number of leaf nodes present in the subtree of the node corresponding to the literal. It was necessary to set multiple binary attributes because our categorical attributes can have a set of values. For example, consider figure 3. If our association rule carried node 138 as an item, a non-empty subset of the seven leaves 140 - 142, 144 - 147 needs to be set. So a random number between 1 and 7 is chosen and random attributes are then set to true.
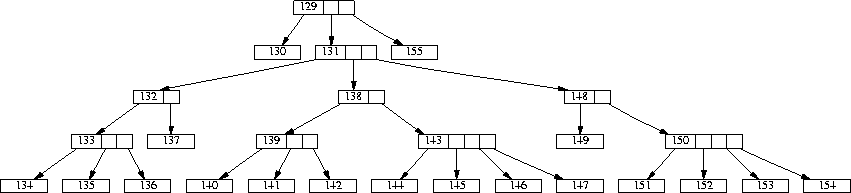
Figure 3: A hierarchy tree including the node number
Here's an example of a short database that was generated. The database after the first randomization looks as follows:
<HIV+ database> 10 elements
0 + 0100000000100000010 1000100000010000001000010000000000010 0 0000000100000000010000000000 0
1 + 0000000000001001010 0000000000000000000000001000000001000 0 0000000000000010000000000010 0
2 + 0000001000000000000 0001000000100000000000000000000001001 0 0000010000000000100010000000 0
3 + 0000000001001000000 0000001000000000000000001001000001010 0 0000000100000000000000100010 0
4 + 0000000010000000000 1000100000000010000000010000000001000 0 0000010000000000100000000100 0
5 + 0000000010100001000 0000000000000000000000000011011001000 0 0000010000000000000100000000 0
6 + 0000000000000000100 0000010000100000000000000000000000000 1 0100000000000000100000000000 0
7 + 0010000000000000000 0000000000000001010010000000001000000 0 0000000010000000100000110100 0
8 + 0000000001101000000 0000100000000000000000000000000000000 0 0000101000000000001000010000 0
9 + 0000001000000000000 0000000000000000000000000000100000010 0 0000010010000000000110101000 0
<HIV- database> 10 elements
0 - 0100000000001000000 1011010000000010000000000000000000000 0 0000000000000000001000000000 0
1 - 0001010000000000000 0000000000000000010000010000000100010 0 0100001000000000000000000001 0
2 - 0101000000000000010 0000000001010000000000001000000000000 0 0000000001000010000000000000 0
3 - 0000100101100001000 0000000000000000000010000001000000000 0 0010000000000000000000000000 0
4 - 0010000000010001000 0000000000000000010100000010000100010 0 0000000010000000000000000000 0
5 - 0000001000000000000 0000000000000000010001000000000010000 0 0000000000000000000100001101 0
6 - 0010000000000000000 0000000100000010000110000000000000000 0 0000000000000100000000000000 0
7 - 0001000010000000000 1000100000001010000000000000000000000 0 0000000000000000000100001000 0
8 - 0000000100100000001 0000000000000000000000000000000000100 0 0000000000000000100000000010 0
9 - 0000000000100010100 0000001000000110000000000000000000000 0 0000000000000000000000000000 0
The tree corresponding to attribute group 0 is shown if figure 4. The data generated looks quite ``dense'': many attributes are set to true. With 96 binary attributes, we expect attributes to be set on average. This is still quite high, which would be against using the square root. But our application is targeted towards a larger application on the order of 1000 attributes -- in which case the average number of set attributes would be around 30, a number which we are anticipating.
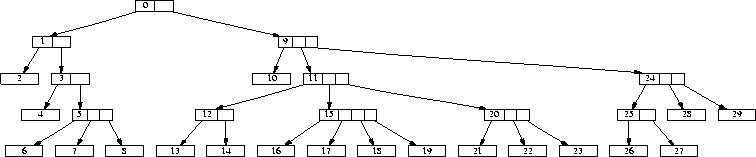
Figure 4: Generated tree for attribute group 0
The association rules were generated in the following steps:
Generating association rule # 0
Association rule 0
Candidate with 4 literals
- 0111100000000000000 0000000000111111111100000000000000000 1 0111111111111111110000000000 0
Positive support: 0, Negative support: 0
Support: 3 Confidence: 0
Generating association rule # 1
Association rule 1
Candidate with 1 literals
- 0000000000000000000 0000000000000000011100000000000000000 0 0000000000000000000000000000 0
Positive support: 2, Negative support: 4
Association rule 1
Candidate with 2 literals
- 0000011111111111111 0000001111000000000000000000000000000 0 0000000000000000000000000000 0
Positive support: 1, Negative support: 2
Association rule 1
Candidate with 1 literals
- 0000000000000000000 0000000000000000000011110000000000000 0 0000000000000000000000000000 0
Positive support: 3, Negative support: 4
Association rule 1
Candidate with 2 literals
- 0000000000000000000 0000000000000000000000000000000000000 1 0000000000000000000000000010 0
Positive support: 0, Negative support: 0
Support: 4 Confidence: 0
Generating association rule # 2
Association rule 2
Candidate with 3 literals
- 0000000000000000000 0000000000000000000011111111000000000 1 0000000000000000000000000000 1
Positive support: 0, Negative support: 0
Support: 7 Confidence: 0
We would like to point out that the association rule number 0 with the pattern 0111100000000000000 for attribute group 0 corresponds to node number 3 in the tree shown in figure 4. During the search for association rule number 2, 3 rules were rejected due to a high match count in the negative tuple set.
The next step was to change the positive and negative tuples to match the support and confidence values of our samples. Remember that support in this case is the number of tuples matching the positive tuple set and the confidence is the number of matching tuples in the negative tuple set.
<HIV+ database> 10 elements
0 + 0100000000100000010 1000100000010000001000010100000000010 1 0000000100000000010000000000 1
1 + 0000000000001001010 0000000000000000000000011100000001000 1 0000000000000010000000000010 1
2 + 0001001000000000000 0001000000111000000010001001000001001 1 0110110000001000100010000000 1
3 + 0000000001001000000 0000001000000000000000001001000001010 0 0000000100000000000000100010 0
4 + 0010000010000000000 1000100000001010100101010000000001000 1 0111011100000100110000000110 1
5 + 0000000010100001000 0000000000000000000011110111011001000 1 0000010000000000000100000000 1
6 + 0000000000000000100 0000010000100000000000010001000000000 1 0100000000000000100000000010 1
7 + 0111100000000000000 0000000000100011110110000000001000000 1 0000000111000100100000110110 0
8 + 0000000001101000000 0000100000000000000000111011000000000 1 0000101000000000001000010010 1
9 + 0000001000000000000 0000000000000000000000000000100000010 0 0000010010000000000110101000 0
<HIV- database> 10 elements
0 - 0100000000001000000 1011010000000010000000000000000000000 0 0000000000000000001000000000 0
1 - 0001010000000000000 0000000000000000010000010000000100010 0 0100001000000000000000000001 0
2 - 0101000000000000010 0000000001010000000000001000000000000 0 0000000001000010000000000000 0
3 - 0000100101100001000 0000000000000000000010000001000000000 0 0010000000000000000000000000 0
4 - 0010000000010001000 0000000000000000010100000010000100010 0 0000000010000000000000000000 0
5 - 0000001000000000000 0000000000000000010001000000000010000 0 0000000000000000000100001101 0
6 - 0010000000000000000 0000000100000010000110000000000000000 0 0000000000000100000000000000 0
7 - 0001000010000000000 1000100000001010000000000000000000000 0 0000000000000000000100001000 0
8 - 0000000100100000001 0000000000000000000000000000000000100 0 0000000000000000100000000010 0
9 - 0000000000100010100 0000001000000110000000000000000000000 0 0000000000000000000000000000 0
We also developed a method to generate a DAG which contains several hierarchy trees. This is very useful because in reality an attribute could belong to different taxonomies.